My Bui (Mimi)
Data Engineer & DataOps
My LinkedIn
My GitHub
Dimensionality Reduction: 1000 fashion MNIST
Goals
1. Try out different Dimensionality Reduction (DR) techniques, and choose the best: TNSE
2. DR for Pre-processing: not very favourable
Data set description
Source: https://github.com/zalandoresearch/fashion-mnist
Fashion-MNIST is a dataset of Zalando’s article images—consisting of a training set of 60,000 examples and a test set of 10,000 examples. Each example is a 28x28 grayscale image, associated with a label from 10 classes. Zalando intends Fashion-MNIST to serve as a direct drop-in replacement for the original MNIST dataset for benchmarking machine learning algorithms. It shares the same image size and structure of training and testing splits.
Each image is 28 pixels in height and 28 pixels in width, for a total of 784 pixels in total. Each pixel has a single pixel-value associated with it, indicating the lightness or darkness of that pixel, with higher numbers meaning darker. This pixel-value is an integer between 0 and 255. The training and test data sets have 785 columns. The first column consists of the class labels (see above), and represents the article of clothing. The rest of the columns contain the pixel-values of the associated image.
Attributes
- 0: T-shirt/top
- 1: Trouser
- 2: Pullover
- 3: Dress
- 4: Coat
- 5: Sandal
- 6: Shirt
- 7: Sneaker
- 8: Bag
- 9: Ankle boot
ML tasks
- Perform Dimentionality Reduction (DR) techniques
- Classification with DR as Pre-processing
We’ll use 1000 rows from the original dataset only.
import pandas as pd
data = pd.read_csv('fashion-mnist.csv')
data
| label | pixel1 | pixel2 | pixel3 | pixel4 | pixel5 | pixel6 | pixel7 | pixel8 | pixel9 | ... | pixel775 | pixel776 | pixel777 | pixel778 | pixel779 | pixel780 | pixel781 | pixel782 | pixel783 | pixel784 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 8 | ... | 103 | 87 | 56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 53 | 99 | ... | 0 | 0 | 0 | 0 | 63 | 53 | 31 | 0 | 0 | 0 |
| 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 137 | 126 | 140 | 0 | 133 | 224 | 222 | 56 | 0 | 0 |
| 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 9995 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 32 | 23 | 14 | 20 | 0 | 0 | 1 | 0 | 0 | 0 |
| 9996 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 2 | 52 | 23 | 28 | 0 | 0 | 0 |
| 9997 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 175 | 172 | 172 | 182 | 199 | 222 | 42 | 0 | 1 | 0 |
| 9998 | 8 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 9999 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 140 | 119 | ... | 111 | 95 | 75 | 44 | 1 | 0 | 0 | 0 | 0 | 0 |
10000 rows × 785 columns
from sklearn.model_selection import train_test_split
X, y = data.iloc[:, 1:].to_numpy(), data.iloc[:, 0].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
class_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
import matplotlib.pyplot as plt
plt.figure(figsize=(10,10))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(X_train[i].reshape(28, 28), cmap="binary")
plt.xlabel(class_names[y_train[i]])
plt.show()

1.1. PCA
# perform PCA and keep 95% of variance
from sklearn.decomposition import PCA
pca = PCA(n_components = 0.95)
X_reduced = pca.fit_transform(X_train)
Most variance lies along the first two principal components
pca.explained_variance_ratio_[:10]
array([0.28701888, 0.18049693, 0.05945456, 0.04980945, 0.03838398,
0.03475959, 0.02353651, 0.01864746, 0.0139102 , 0.01324683])
# reconstruct data from compressed data
pca = PCA(n_components = 179)
X_reduced = pca.fit_transform(X_train)
X_recovered = pca.inverse_transform(X_reduced)
import matplotlib as mpl
import matplotlib.pyplot as plt
print('non-compressed image')
plt.imshow(X_train[100].reshape(28, 28), cmap="binary")
plt.axis("off")
plt.show()
non-compressed image

print('95% variance compressed image')
plt.imshow(X_recovered[100].reshape(28, 28), cmap="binary")
plt.axis("off")
plt.show()
95% variance compressed image

# helper function for plotting
from sklearn.preprocessing import MinMaxScaler
from matplotlib.offsetbox import AnnotationBbox, OffsetImage
import numpy as np
def plot_digits(X, y, min_distance=0.05, images=None, figsize=(13, 10)):
# Let's scale the input features so that they range from 0 to 1
X_normalized = MinMaxScaler().fit_transform(X)
# Now we create the list of coordinates of the digits plotted so far.
# We pretend that one is already plotted far away at the start, to
# avoid `if` statements in the loop below
neighbors = np.array([[10., 10.]])
# The rest should be self-explanatory
plt.figure(figsize=figsize)
cmap = mpl.cm.get_cmap("prism")
digits = np.unique(y)
for digit in digits:
plt.scatter(X_normalized[y == digit, 0], X_normalized[y == digit, 1], c=[cmap(digit / 9)])
plt.axis("off")
ax = plt.gcf().gca() # get current axes in current figure
for index, image_coord in enumerate(X_normalized):
closest_distance = np.linalg.norm(np.array(neighbors) - image_coord, axis=1).min()
if closest_distance > min_distance:
neighbors = np.r_[neighbors, [image_coord]]
if images is None:
plt.text(image_coord[0], image_coord[1], str(int(y[index])),
color=cmap(y[index] / 9), fontdict={"weight": "bold", "size": 16})
else:
image = images[index].reshape(28, 28)
imagebox = AnnotationBbox(OffsetImage(image, cmap="binary"), image_coord)
ax.add_artist(imagebox)
X_pca_reduced = PCA(n_components=2, random_state=42).fit_transform(X_train)
plot_digits(X_pca_reduced, y_train)
plt.show()
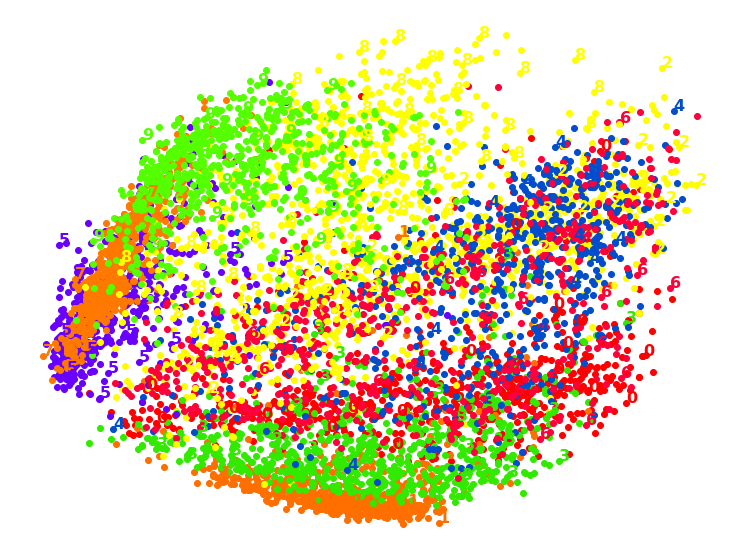
1.2. LDA
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
X_lda_reduced = LinearDiscriminantAnalysis(n_components=2).fit_transform(X_train, y_train)
plot_digits(X_lda_reduced, y_train, figsize=(12,12))
plt.show()
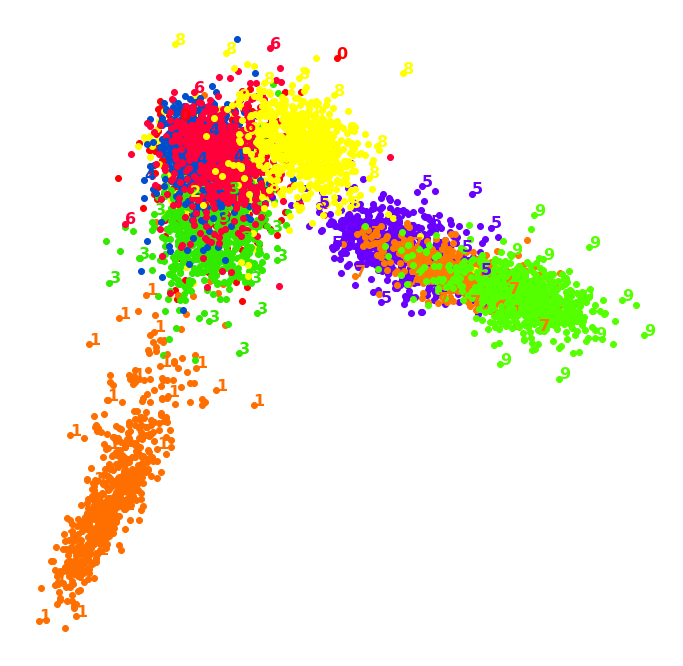
1.3. LLE
from sklearn.pipeline import Pipeline
from sklearn.manifold import LocallyLinearEmbedding
pca_lle = Pipeline([
("pca", PCA(n_components=0.95, random_state=42)),
("lle", LocallyLinearEmbedding(n_components=2, random_state=42)),
])
X_pca_lle_reduced = pca_lle.fit_transform(X_train)
plot_digits(X_pca_lle_reduced, y_train)
plt.show()
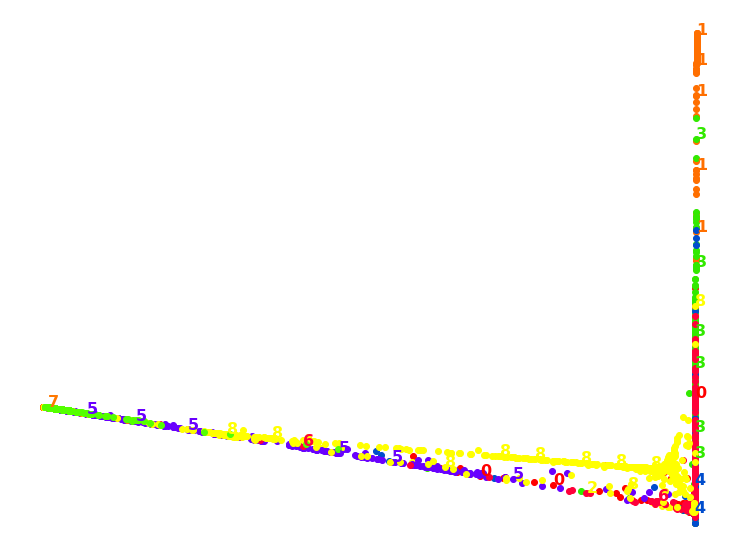
1.4. MDS
from sklearn.manifold import MDS
pca_mds = Pipeline([
("pca", PCA(n_components=0.95, random_state=42)),
("mds", MDS(n_components=2, random_state=42)),
])
X_pca_mds_reduced = pca_mds.fit_transform(X_train[:1000])
plot_digits(X_pca_mds_reduced, y_train[:1000])
plt.show()
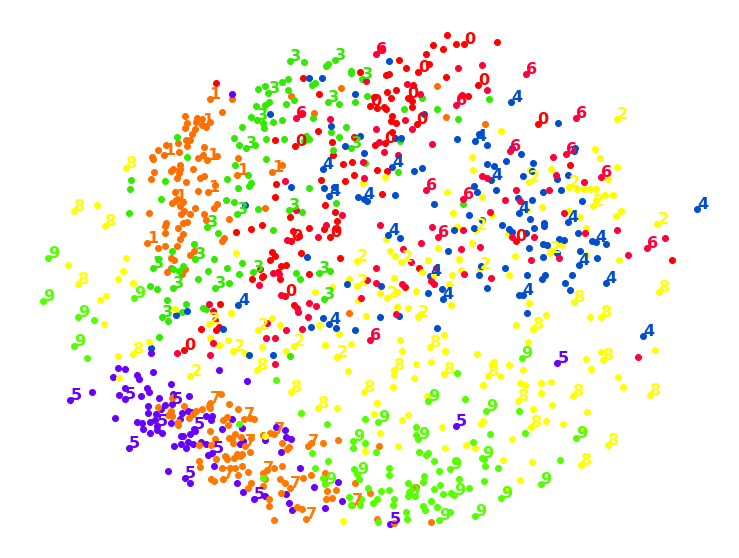
1.5. Isomap
from sklearn.manifold import Isomap
pca_isomap = Pipeline([
("pca", PCA(n_components=0.95, random_state=42)),
("isomap", Isomap(n_components=2)),
])
X_pca_isomap_reduced = pca_isomap.fit_transform(X_train)
plot_digits(X_pca_isomap_reduced, y_train)
plt.show()
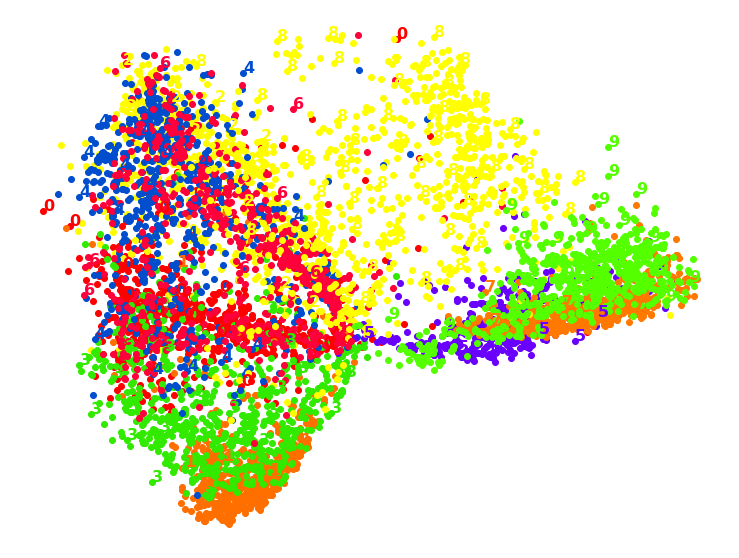
1.6. TSNE
from sklearn.manifold import TSNE
pca_tsne = Pipeline([
("pca", PCA(n_components=0.95, random_state=42)),
("tsne", TSNE(n_components=2, random_state=42)),
])
X_pca_tsne_reduced = pca_tsne.fit_transform(X_train)
plot_digits(X_pca_tsne_reduced, y_train)
plt.show()
TSNS gives the clearest distinction between classes.
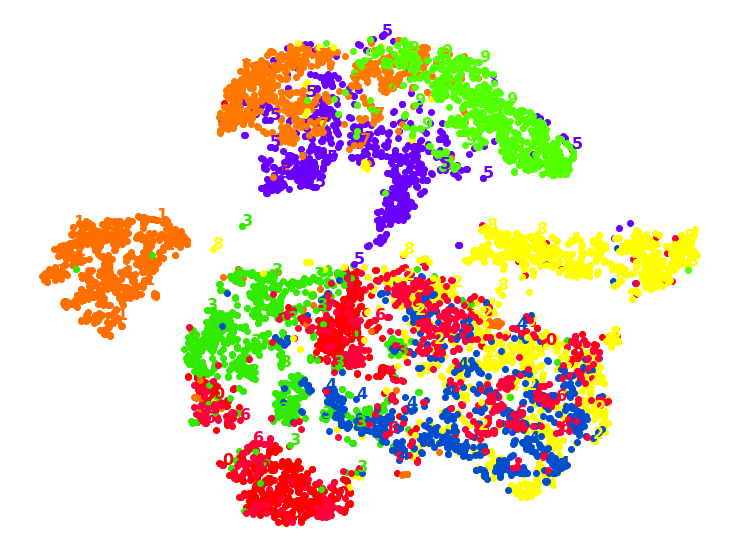
1.7. TSNE with ‘rbf’ KPCA
from sklearn.decomposition import KernelPCA
from sklearn.manifold import TSNE
k_pca_tsne = Pipeline([
("kpca", KernelPCA(n_components=2, kernel='rbf', random_state=42)),
("tsne", TSNE(n_components=2, random_state=42)),
])
X_pca_tsne_reduced = k_pca_tsne.fit_transform(X_train)
plot_digits(X_pca_tsne_reduced, y_train)
plt.show()
1.8. TSNE with ‘poly’ KPCA
k_pca_tsne = Pipeline([
("kpca", KernelPCA(n_components=2, kernel='poly', random_state=42)),
("tsne", TSNE(n_components=2, random_state=42)),
])
X_pca_tsne_reduced = k_pca_tsne.fit_transform(X_train)
plot_digits(X_pca_tsne_reduced, y_train)
plt.show()
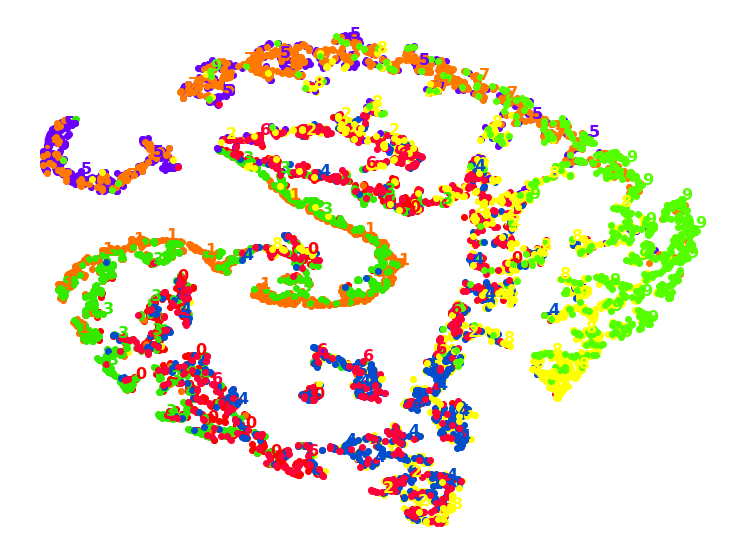
1.9. TSNE with ‘sigmoid’ PCA
k_pca_tsne = Pipeline([
("kpca", KernelPCA(n_components=2, kernel='sigmoid', random_state=42)),
("tsne", TSNE(n_components=2, random_state=42)),
])
X_pca_tsne_reduced = k_pca_tsne.fit_transform(X_train)
plot_digits(X_pca_tsne_reduced, y_train)
plt.show()

2.1. Classification with PCA and TNSE
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression()
lr.fit(X_pca_tsne_reduced, y_train)
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=100,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=None, solver='lbfgs', tol=0.0001, verbose=0,
warm_start=False)
pca_tsne = Pipeline([
("pca", PCA(n_components=0.95, random_state=42)),
("tsne", TSNE(n_components=2, random_state=42)),
])
X_pca_tsne_reduced_test = pca_tsne.fit_transform(X_test)
from sklearn.metrics import accuracy_score
accuracy_score(y_test, lr.predict(X_pca_tsne_reduced_test))
0.0048484848484848485
The result is very bad. So we can try simple PCA with 2 components.
lr = LogisticRegression(max_iter=100000)
lr.fit(X_pca_reduced, y_train)
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=100000,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=None, solver='lbfgs', tol=0.0001, verbose=0,
warm_start=False)
X_pca_reduced_test = PCA(n_components=2, random_state=42).fit_transform(X_test)
accuracy_score(y_test, lr.predict(X_pca_reduced_test))
0.5130303030303031