My Bui (Mimi)
Data Engineer & DataOps
My LinkedIn
My GitHub
Supervised learning: regression and classification of math final grades
Goals
1. Regression - final grade: MSE = 6.1558
2. Binary classification – pass if ≥ 10 else fail: 9 mis-classified points
3. 5-Level classification – based on the Erasmus grading: 7 mis-classified points
Attributes
- school - student’s school (binary: ‘GP’ - Gabriel Pereira or ‘MS’ - Mousinho da Silveira)
- sex - student’s sex (binary: ‘F’ - female or ‘M’ - male)
- age - student’s age (numeric: from 15 to 22)
- address - student’s home address type (binary: ‘U’ - urban or ‘R’ - rural)
- famsize - family size (binary: ‘LE3’ - less or equal to 3 or ‘GT3’ - greater than 3)
- Pstatus - parent’s cohabitation status (binary: ‘T’ - living together or ‘A’ - apart)
- Medu - mother’s education (numeric: 0 - none, 1 - primary education (4th grade), 2 - 5th to 9th grade, 3 - secondary education or 4 - higher education)
- Fedu - father’s education (numeric: 0 - none, 1 - primary education (4th grade), 2 - 5th to 9th grade, 3 - secondary education or 4 - higher education)
- Mjob - mother’s job (nominal: ‘teacher’, ‘health’ care related, civil ‘services’ (e.g. administrative or police), ‘at_home’ or ‘other’)
- Fjob - father’s job (nominal: ‘teacher’, ‘health’ care related, civil ‘services’ (e.g. administrative or police), ‘at_home’ or ‘other’)
- reason - reason to choose this school (nominal: close to ‘home’, school ‘reputation’, ‘course’ preference or ‘other’)
- guardian - student’s guardian (nominal: ‘mother’, ‘father’ or ‘other’)
- traveltime - home to school travel time (numeric: 1 - <15 min., 2 - 15 to 30 min., 3 - 30 min. to 1 hour, or 4 - >1 hour)
- studytime - weekly study time (numeric: 1 - <2 hours, 2 - 2 to 5 hours, 3 - 5 to 10 hours, or 4 - >10 hours)
- failures - number of past class failures (numeric: n if 1<=n<3, else 4)
- schoolsup - extra educational support (binary: yes or no)
- famsup - family educational support (binary: yes or no)
- paid - extra paid classes within the course subject (Math or Portuguese) (binary: yes or no)
- activities - extra-curricular activities (binary: yes or no)
- nursery - attended nursery school (binary: yes or no)
- higher - wants to take higher education (binary: yes or no)
- internet - Internet access at home (binary: yes or no)
- romantic - with a romantic relationship (binary: yes or no)
- famrel - quality of family relationships (numeric: from 1 - very bad to 5 - excellent)
- freetime - free time after school (numeric: from 1 - very low to 5 - very high)
- goout - going out with friends (numeric: from 1 - very low to 5 - very high)
- Dalc - workday alcohol consumption (numeric: from 1 - very low to 5 - very high)
- Walc - weekend alcohol consumption (numeric: from 1 - very low to 5 - very high)
- health - current health status (numeric: from 1 - very bad to 5 - very good)
- absences - number of school absences (numeric: from 0 to 93)
- G1 - first period grade (numeric: from 0 to 20)
- G2 - second period grade (numeric: from 0 to 20)
- G3 - final grade (numeric: from 0 to 20, output target)
ML tasks
- Regression - final grade G3
- Binary classification – pass if G3 ≥ 10 else fail
- 5-Level classification – based on the Erasmus grade conversion system
import pandas as pd
import numpy as np
from sklearn.linear_model import Ridge
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso
data = pd.read_csv('student/student-mat.csv', sep=';')
data
| school | sex | age | address | famsize | Pstatus | Medu | Fedu | Mjob | Fjob | ... | famrel | freetime | goout | Dalc | Walc | health | absences | G1 | G2 | G3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GP | F | 18 | U | GT3 | A | 4 | 4 | at_home | teacher | ... | 4 | 3 | 4 | 1 | 1 | 3 | 6 | 5 | 6 | 6 |
| 1 | GP | F | 17 | U | GT3 | T | 1 | 1 | at_home | other | ... | 5 | 3 | 3 | 1 | 1 | 3 | 4 | 5 | 5 | 6 |
| 2 | GP | F | 15 | U | LE3 | T | 1 | 1 | at_home | other | ... | 4 | 3 | 2 | 2 | 3 | 3 | 10 | 7 | 8 | 10 |
| 3 | GP | F | 15 | U | GT3 | T | 4 | 2 | health | services | ... | 3 | 2 | 2 | 1 | 1 | 5 | 2 | 15 | 14 | 15 |
| 4 | GP | F | 16 | U | GT3 | T | 3 | 3 | other | other | ... | 4 | 3 | 2 | 1 | 2 | 5 | 4 | 6 | 10 | 10 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 390 | MS | M | 20 | U | LE3 | A | 2 | 2 | services | services | ... | 5 | 5 | 4 | 4 | 5 | 4 | 11 | 9 | 9 | 9 |
| 391 | MS | M | 17 | U | LE3 | T | 3 | 1 | services | services | ... | 2 | 4 | 5 | 3 | 4 | 2 | 3 | 14 | 16 | 16 |
| 392 | MS | M | 21 | R | GT3 | T | 1 | 1 | other | other | ... | 5 | 5 | 3 | 3 | 3 | 3 | 3 | 10 | 8 | 7 |
| 393 | MS | M | 18 | R | LE3 | T | 3 | 2 | services | other | ... | 4 | 4 | 1 | 3 | 4 | 5 | 0 | 11 | 12 | 10 |
| 394 | MS | M | 19 | U | LE3 | T | 1 | 1 | other | at_home | ... | 3 | 2 | 3 | 3 | 3 | 5 | 5 | 8 | 9 | 9 |
395 rows × 33 columns
Feature Engineering
o = list(data.select_dtypes(include='object').columns)
o
['school',
'sex',
'address',
'famsize',
'Pstatus',
'Mjob',
'Fjob',
'reason',
'guardian',
'schoolsup',
'famsup',
'paid',
'activities',
'nursery',
'higher',
'internet',
'romantic']
num_data = data.copy().drop(o, axis=1)
txt_data = data.loc[:, o]
# categorize these nominal features and make corresponding dummies
for i in o:
txt_data[i] = data[i].astype('category')
txt_data = pd.concat([txt_data,
pd.get_dummies(txt_data.select_dtypes(include=['category']))], axis=1).drop(i, axis=1)
trans_data = pd.concat([txt_data, num_data], axis=1)
trans_data
| school_GP | school_MS | sex_F | sex_M | address_R | address_U | famsize_GT3 | famsize_LE3 | Pstatus_A | Pstatus_T | ... | famrel | freetime | goout | Dalc | Walc | health | absences | G1 | G2 | G3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | ... | 4 | 3 | 4 | 1 | 1 | 3 | 6 | 5 | 6 | 6 |
| 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | ... | 5 | 3 | 3 | 1 | 1 | 3 | 4 | 5 | 5 | 6 |
| 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 4 | 3 | 2 | 2 | 3 | 3 | 10 | 7 | 8 | 10 |
| 3 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | ... | 3 | 2 | 2 | 1 | 1 | 5 | 2 | 15 | 14 | 15 |
| 4 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | ... | 4 | 3 | 2 | 1 | 2 | 5 | 4 | 6 | 10 | 10 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 390 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | ... | 5 | 5 | 4 | 4 | 5 | 4 | 11 | 9 | 9 | 9 |
| 391 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 2 | 4 | 5 | 3 | 4 | 2 | 3 | 14 | 16 | 16 |
| 392 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | ... | 5 | 5 | 3 | 3 | 3 | 3 | 3 | 10 | 8 | 7 |
| 393 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | ... | 4 | 4 | 1 | 3 | 4 | 5 | 0 | 11 | 12 | 10 |
| 394 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 3 | 2 | 3 | 3 | 3 | 5 | 5 | 8 | 9 | 9 |
395 rows × 59 columns
1. Linear Regression (LR)
X = trans_data.iloc[:, :58].values
y = trans_data.iloc[:, 58].values
X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True, random_state=0)
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
mse = mean_squared_error(y_test, y_pred)
mse
6.14478692382273
Regularization with Ridge Regression (RR) which sets some features to be close to zero
Try out different values of alpha for regularization
ridge_df = pd.DataFrame()
ridge_mse = []
for alpha in np.arange(1, 200, 1):
ridge = Ridge(alpha=alpha)
ridge.fit(X_train, y_train)
ridge_df[alpha] = ridge.coef_
ridge_mse.append(mean_squared_error(ridge.predict(X_test), y_test))
ridge_df = ridge_df.T
ridge_df
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | -0.175165 | 0.175165 | -0.103920 | 0.103920 | -0.081354 | 0.081354 | -0.056786 | 0.056786 | 0.145694 | -0.145694 | ... | -0.157074 | 0.158110 | 0.042826 | -0.061411 | -0.212318 | 0.233830 | 0.094764 | 0.053072 | 0.155966 | 0.952054 |
| 2 | -0.172091 | 0.172091 | -0.101828 | 0.101828 | -0.079433 | 0.079433 | -0.056676 | 0.056676 | 0.142775 | -0.142775 | ... | -0.157279 | 0.157483 | 0.042661 | -0.061260 | -0.208337 | 0.231553 | 0.094180 | 0.052949 | 0.155394 | 0.952180 |
| 3 | -0.169075 | 0.169075 | -0.099830 | 0.099830 | -0.077643 | 0.077643 | -0.056511 | 0.056511 | 0.139965 | -0.139965 | ... | -0.157385 | 0.156832 | 0.042499 | -0.061107 | -0.204515 | 0.229363 | 0.093608 | 0.052826 | 0.154913 | 0.952247 |
| 4 | -0.166125 | 0.166125 | -0.097920 | 0.097920 | -0.075970 | 0.075970 | -0.056303 | 0.056303 | 0.137262 | -0.137262 | ... | -0.157407 | 0.156162 | 0.042341 | -0.060955 | -0.200841 | 0.227253 | 0.093050 | 0.052703 | 0.154511 | 0.952261 |
| 5 | -0.163244 | 0.163244 | -0.096091 | 0.096091 | -0.074401 | 0.074401 | -0.056062 | 0.056062 | 0.134660 | -0.134660 | ... | -0.157361 | 0.155477 | 0.042186 | -0.060803 | -0.197303 | 0.225217 | 0.092506 | 0.052580 | 0.154178 | 0.952231 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 195 | -0.030416 | 0.030416 | -0.029442 | 0.029442 | -0.030304 | 0.030304 | -0.023686 | 0.023686 | 0.032109 | -0.032109 | ... | -0.098194 | 0.072108 | 0.031251 | -0.043248 | -0.037198 | 0.112158 | 0.055302 | 0.042920 | 0.213843 | 0.863639 |
| 196 | -0.030254 | 0.030254 | -0.029375 | 0.029375 | -0.030265 | 0.030265 | -0.023620 | 0.023620 | 0.031990 | -0.031990 | ... | -0.098020 | 0.071881 | 0.031214 | -0.043190 | -0.036993 | 0.111923 | 0.055201 | 0.042896 | 0.214141 | 0.863209 |
| 197 | -0.030093 | 0.030093 | -0.029308 | 0.029308 | -0.030227 | 0.030227 | -0.023555 | 0.023555 | 0.031871 | -0.031871 | ... | -0.097846 | 0.071656 | 0.031176 | -0.043131 | -0.036789 | 0.111688 | 0.055100 | 0.042872 | 0.214438 | 0.862781 |
| 198 | -0.029933 | 0.029933 | -0.029242 | 0.029242 | -0.030189 | 0.030189 | -0.023490 | 0.023490 | 0.031754 | -0.031754 | ... | -0.097674 | 0.071432 | 0.031139 | -0.043073 | -0.036587 | 0.111456 | 0.055000 | 0.042848 | 0.214734 | 0.862353 |
| 199 | -0.029774 | 0.029774 | -0.029177 | 0.029177 | -0.030152 | 0.030152 | -0.023426 | 0.023426 | 0.031638 | -0.031638 | ... | -0.097502 | 0.071209 | 0.031102 | -0.043015 | -0.036387 | 0.111224 | 0.054900 | 0.042824 | 0.215029 | 0.861926 |
199 rows × 58 columns
Among the last 10 features, G2 is given the highest importance, and secondly G1.
fig, ax = plt.subplots(figsize=(10, 5))
ax.plot(ridge_df.iloc[:, 50:])
ax.axhline(y=0, color='black', linestyle='--')
ax.set_xlabel("alpha")
ax.set_ylabel("coeficients")
ax.set_title("RR for the last 10 features")
ax.legend(labels=list(trans_data.iloc[:, 50:-1].columns))
ax.grid(True)
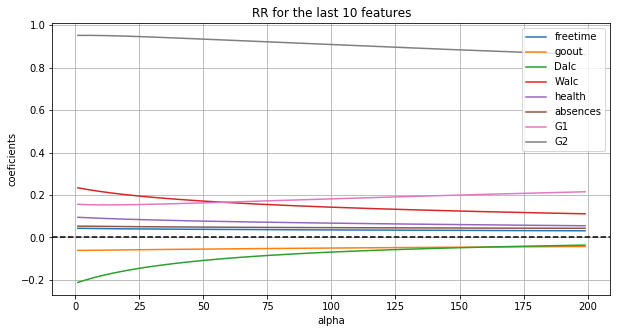
The plot of MSE shows the optimum alpha is in [70, 80].
fig, ax = plt.subplots(figsize=(10, 5))
plt.plot(ridge_mse)
ax.set_xlabel("alpha")
ax.set_ylabel("mse")
ax.set_title("RR MSE trace")
ax.axhline(y=mse, color='red', linestyle='--')
ax.legend(['RR', 'OLS'])
plt.show()
Regularization with Lasso which ignores some features completely
Try out different values of alpha for regularization. The model uses more features when alpha is smaller.
lasso_df = pd.DataFrame()
lasso_mse = []
a_range = list(reversed([1, 0.1, 0.01, 0.001, 0.0001, 0.00001, 0.000001]))
for alpha in a_range:
lasso = Lasso(alpha=alpha, max_iter=100000)
lasso.fit(X_train, y_train)
print(lasso.coef_)
lasso_df[alpha] = lasso.coef_
lasso_mse.append(mean_squared_error(lasso.predict(X_test), y_test))
lasso_df = lasso_df.T
lasso_df
fig, ax = plt.subplots(figsize=(10, 5))
ax.plot(lasso_df.iloc[:, 50:])
ax.axhline(y=0, color='black', linestyle='--')
ax.set_xlabel("alpha")
ax.set_ylabel("coeficients")
ax.set_title("Lasso for the last 10 features")
labels = [item.get_text() for item in ax.get_xticklabels()]
for i in range(0, len(labels)-1):
labels[i] = a_range[i]
ax.set_xticklabels(labels, rotation=45)
ax.legend(labels=list(trans_data.iloc[:, 50:-1].columns))
ax.grid(True)
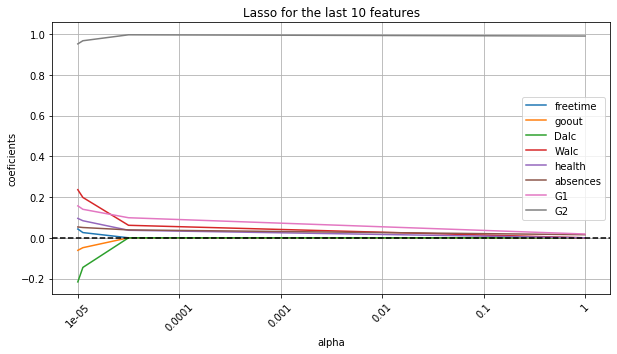
At alpha = 0.01, the model achieves the best performance which uses 42 features.
fig, ax = plt.subplots(figsize=(10, 5))
plt.plot(lasso_mse)
ax.set_xlabel("alpha")
ax.set_ylabel("mse")
ax.set_title("Lasso MSE trace")
ax.axhline(y=mse, color='red', linestyle='--')
labels = [item.get_text() for item in ax.get_xticklabels()]
for i in range(0, len(labels)-2):
labels[i+1] = a_range[i]
ax.set_xticklabels(labels, rotation=45)
ax.legend(['Lasso', 'OLS'])
plt.show()
a_001 = lasso_df.iloc[4, :]
len(a_001[a_001 != 0])
42
Robust Regression (RbR) for outliers
# make some strong outliers for X
X_errors = X.copy()
X_errors[::3] = 100
from sklearn.model_selection import train_test_split
X_train_error, X_test_error, y_train, y_test = train_test_split(X_errors, y, shuffle=True, random_state=0)
from sklearn.linear_model import (LinearRegression, TheilSenRegressor, RANSACRegressor, HuberRegressor)
from sklearn.metrics import mean_squared_error
estimators = [('OLS', LinearRegression()),
('Theil-Sen', TheilSenRegressor(random_state=42, max_iter=10000)),
('RANSAC', RANSACRegressor(random_state=42, max_trials=10000)),
('HuberRegressor', HuberRegressor(max_iter=10000))]
TheilSen is good for strong outliers, both in direction X and y.
for e in estimators:
m = e[1].fit(X_train_error, y_train)
print("For " + e[0] + " technique:")
print("Training set score: {:.2f}".format(m.score(X_train_error, y_train)))
print("Test set score: {:.2f}".format(m.score(X_test_error, y_test)))
print("MSE: {:.2f} \n".format(mean_squared_error(y_test, m.predict(X_test_error))))
For OLS technique:
Training set score: 0.66
Test set score: 0.47
MSE: 15.04
For Theil-Sen technique:
Training set score: 0.66
Test set score: 0.48
MSE: 14.73
For RANSAC technique:
Training set score: 0.61
Test set score: 0.43
MSE: 16.09
For HuberRegressor technique:
Training set score: 0.64
Test set score: 0.46
MSE: 15.39
# make some strong outliers for y
y_errors = y.copy()
y_errors[::2] = 100
from sklearn.model_selection import train_test_split
X_train, X_test, y_train_error, y_test_error = train_test_split(X, y_errors, shuffle=True, random_state=0)
for e in estimators:
m = e[1].fit(X_train, y_train_error)
print("For " + e[0] + " technique:")
print("Training set score: {:.2f}".format(m.score(X_train, y_train_error)))
print("Test set score: {:.2f}".format(m.score(X_test, y_test_error)))
print("MSE: {:.2f} \n".format(mean_squared_error(y_test_error, m.predict(X_test))))
For OLS technique:
Training set score: 0.10
Test set score: -0.25
MSE: 2508.31
For Theil-Sen technique:
Training set score: 0.09
Test set score: -0.27
MSE: 2547.76
For RANSAC technique:
Training set score: -1.51
Test set score: -2.12
MSE: 6232.98
For HuberRegressor technique:
Training set score: 0.10
Test set score: -0.30
MSE: 2592.34
Polynomial Regression with features G1 & G2 to predict G3
X = trans_data.iloc[:, 56:58].values
y = trans_data.iloc[:, 58].values
from sklearn.preprocessing import PolynomialFeatures
pol_mse = []
d_range = np.arange(2, 8, 1)
for d in d_range:
pol = PolynomialFeatures(degree=d)
pol.fit(X_train, y_train)
X_2 = pol.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(X_2, y, random_state=0)
# OLS
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
pol_mse.append(mean_squared_error(y_test, y_pred))
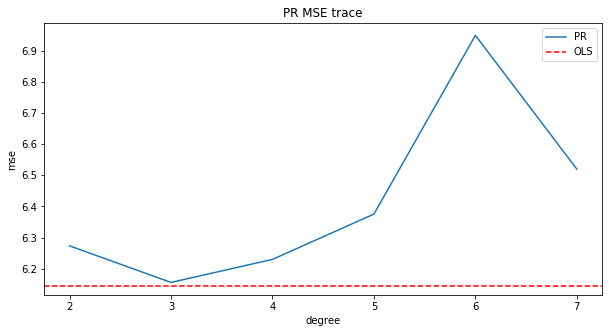
pol_mse
[6.273301123930405,
6.155807238512367,
6.229853804336613,
6.375376965526311,
6.949451552084533,
6.519870726895505]
Polynomial Regression with 2 features so far performs the best compared to other models
fig, ax = plt.subplots(figsize=(10, 5))
plt.plot(pol_mse)
ax.set_xlabel("degree")
ax.set_ylabel("mse")
ax.set_title("PR MSE trace")
ax.axhline(y=mse, color='red', linestyle='--')
ax.set_xticklabels(np.arange(1, 8, 1))
ax.legend(['PR', 'OLS'])
plt.show()
2. Binary classification for G3
Create a new column that categorizes the feature G3 as fail if < 10, and pass otherwise
def pass_grade(x):
if x <= 10:
return 0
else:
return 1
trans_data['pass'] = trans_data['G3'].apply(lambda x: pass_grade(x))
trans_data[['G3', 'pass']]
| G3 | pass | |
|---|---|---|
| 0 | 6 | 0 |
| 1 | 6 | 0 |
| 2 | 10 | 0 |
| 3 | 15 | 1 |
| 4 | 10 | 0 |
| ... | ... | ... |
| 390 | 9 | 0 |
| 391 | 16 | 1 |
| 392 | 7 | 0 |
| 393 | 10 | 0 |
| 394 | 9 | 0 |
395 rows × 2 columns
from sklearn.svm import LinearSVC
lsvc = LinearSVC(max_iter=100000).fit(X_train, y_train)
print("Training set score: {:.3f}".format(lsvc.score(X_train, y_train)))
print("Test set score: {:.3f}".format(lsvc.score(X_test, y_test)))
Training set score: 0.986
Test set score: 0.899
X = trans_data.iloc[:, :58].values
y = trans_data['pass'].values
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
The model performance is best acheived when c is from 3 to 9, with 7 type-1 faults and 1 type-2 faults.
for c in list(range(1, 11)) + [100]:
logReg = LogisticRegression(C=c, max_iter=1000000)
logReg.fit(X_train, y_train)
print("c is " + str(c))
print(confusion_matrix(y_test, logReg.predict(X_test)))
print('\n')
c is 1
[[46 5]
[ 0 48]]
c is 2
[[46 5]
[ 1 47]]
c is 3
[[44 7]
[ 1 47]]
c is 4
[[44 7]
[ 1 47]]
c is 5
[[44 7]
[ 1 47]]
c is 6
[[44 7]
[ 1 47]]
c is 7
[[44 7]
[ 1 47]]
c is 8
[[44 7]
[ 1 47]]
c is 9
[[44 7]
[ 1 47]]
c is 10
[[43 8]
[ 1 47]]
c is 100
[[43 8]
[ 1 47]]
3. Multi-value classification for G3
Create a new column that categorizes the feature G3 according to ERASMUS scale
def pass_grade_5(x):
if 0 <= x <= 9:
return 5
elif 10 <= x <= 11:
return 4
elif 12 <= x <= 13:
return 3
elif 14 <= x <= 15:
return 2
else:
return 1
trans_data['pass_5'] = trans_data['G3'].apply(lambda x: pass_grade_5(x))
trans_data
| school_GP | school_MS | sex_F | sex_M | address_R | address_U | famsize_GT3 | famsize_LE3 | Pstatus_A | Pstatus_T | ... | goout | Dalc | Walc | health | absences | G1 | G2 | G3 | pass | pass_5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | ... | 4 | 1 | 1 | 3 | 6 | 5 | 6 | 6 | 0 | 5 |
| 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | ... | 3 | 1 | 1 | 3 | 4 | 5 | 5 | 6 | 0 | 5 |
| 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 2 | 2 | 3 | 3 | 10 | 7 | 8 | 10 | 0 | 4 |
| 3 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | ... | 2 | 1 | 1 | 5 | 2 | 15 | 14 | 15 | 1 | 2 |
| 4 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | ... | 2 | 1 | 2 | 5 | 4 | 6 | 10 | 10 | 0 | 4 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 390 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | ... | 4 | 4 | 5 | 4 | 11 | 9 | 9 | 9 | 0 | 5 |
| 391 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 5 | 3 | 4 | 2 | 3 | 14 | 16 | 16 | 1 | 1 |
| 392 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | ... | 3 | 3 | 3 | 3 | 3 | 10 | 8 | 7 | 0 | 5 |
| 393 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | ... | 1 | 3 | 4 | 5 | 0 | 11 | 12 | 10 | 0 | 4 |
| 394 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 3 | 3 | 3 | 5 | 5 | 8 | 9 | 9 | 0 | 5 |
395 rows × 61 columns
X = trans_data.iloc[:, :58].values
y = trans_data['pass_5'].values
The model performance is best acheived when c is >= 8, with 6 type-1 faults and 1 type-2 faults.
for c in list(range(1, 11)) + [100]:
lsvc = LinearSVC(C=c, max_iter=1000000)
lsvc.fit(X_train, y_train)
print("c is " + str(c))
print(confusion_matrix(y_test, lsvc.predict(X_test)))
print('\n')
c is 1
[[43 8]
[ 2 46]]
c is 2
[[43 8]
[ 2 46]]
c is 3
[[44 7]
[ 2 46]]
c is 4
[[44 7]
[ 1 47]]
c is 5
[[44 7]
[ 1 47]]
c is 6
[[44 7]
[ 1 47]]
c is 7
[[44 7]
[ 1 47]]
c is 8
[[45 6]
[ 1 47]]
c is 9
[[45 6]
[ 1 47]]
c is 10
[[45 6]
[ 1 47]]
c is 100
[[45 6]
[ 1 47]]